Submitting Raw Data to EGA
As part of the EGA submission process, EGA requires metadata in order to associate samples with the raw files you uploaded. This metadata is crucial for users to map your DCC-submitted samples to raw data files at EGA. The following instructions are meant to provide ICGC members with guidance on submitting metadata to the European Genome-Phenome Archive (EGA). ICGC members are encouraged to consult the EGA submission guidelines prior to data submission.
Overview of the EGA metadata XML files
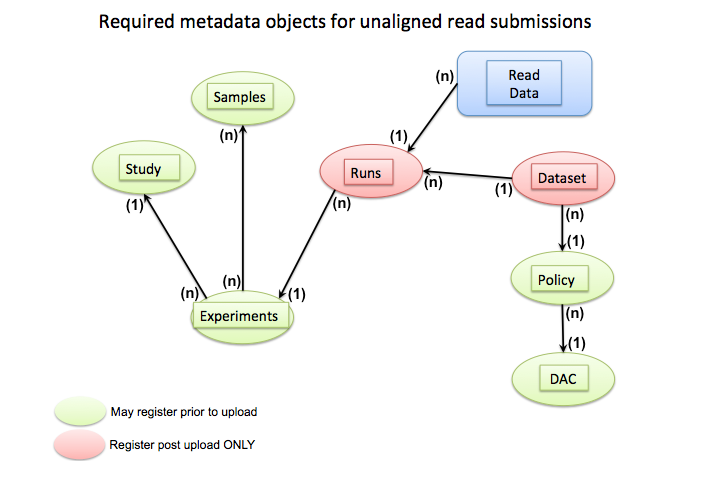
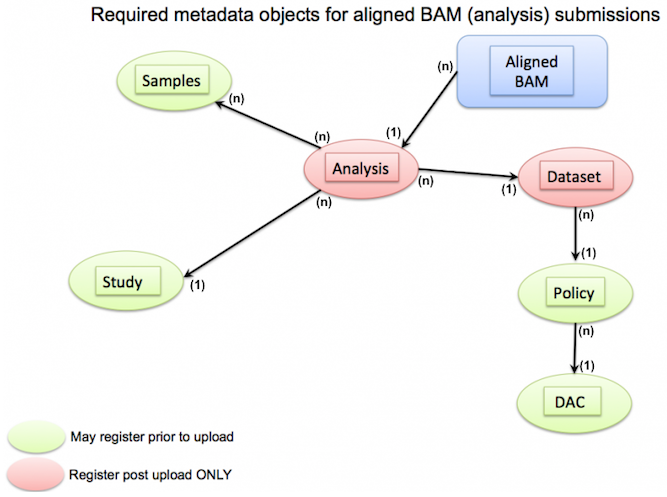
Below are example template files for POLICY, STUDY, DATASET, SAMPLE, EXPERIMENT, and RUN metadata.
Submissions from ICGC members do not need to include a dac.xml, as the ICGC DAC (Data Access Committee) already exists in the EGA system. The submitters only need to refer to the ICGC DAC in the policy.xml, as shown in the following policy.xml example.
POLICY XML file
<?xml version = '1.0' encoding = 'UTF-8'?>
<POLICY_SET>
<POLICY alias="ICGC Data Access Agreements" center_name="YOUR_CENTER_NAME_HERE. See http://docs.icgc.org/submission/institutions/ (eg. OICR)" broker_name="EGA">
<TITLE>ICGC Data Access</TITLE>
<DAC_REF refname="ICGC Cancer Genome Projects" refcenter="ICGC"/>
<DAC_REF accession="EGAC00001000010"/>
<POLICY_TEXT>Please use the ICGC website for applying access to the data</POLICY_TEXT>
<POLICY_LINKS>
<POLICY_LINK>
<URL_LINK>
<LABEL>ICGC Data Access Agreements</LABEL>
<URL>http://www.icgc.org</URL>
</URL_LINK>
</POLICY_LINK>
</POLICY_LINKS>
</POLICY>
</POLICY_SET>
- Please note in the above example that submitting centers must populate their own center name in the center_name attribute of the POLICY element. Please use your center name as specified in List of Institutions at http://docs.icgc.org/submission/institutions/. If your center’s name is incorrect or missing from here, please contact the DCC to have your center’s record added or revised.
- The value of center_name should reflect the submitter’s affiliation and should remain consistent in the metadata XML for all subsequent submissions to EGA.
DATASET and STUDY XML files
The following examples of DATASET and STUDY xml files are written as per EGA's specifications with key items required for all ICGC submissions populated with examples.
DATASET XML file
<?xml version = '1.0' encoding = 'UTF-8'?>
<DATASETS>
<DATASET alias="TODO:UNIQUE NAME e.g. DS20110311" center_name="TODO: ACCOUNT CENTER_NAME ACRONYM e.g. OICR" broker_name="EGA">
<TITLE>TODO:DATASET TITLE e.g. Control samples for X</TITLE>
<DATASET_TYPE>TODO: CONTROLLED TERMS END OF XML e.g. Whole genome sequencing </DATASET_TYPE>
<RUN_REF accession="TODO:ACCESSION NUMBER(EGAR)"/>
<RUN_REF accession="TODO:ACCESSION NUMBER(EGAR)"/>
<ANALYSIS_REF accession="TO DO: ACCESSION NUMBER (EGAZ)"/>
<!--
EGAR/EGAZ accession numbers are provided upon the receipt of the submission of
an Run/analysis XML.
-->
<POLICY_REF accession="EGAP00001000011" refcenter="ICGC"/>
<DATASET_LINKS>
<DATASET_LINK>
<URL_LINK>
<LABEL>ICGC Data Portal</LABEL>
<URL>http://dcc.icgc.org</URL>
</URL_LINK>
</DATASET_LINK>
</DATASET_LINKS>
<DATASET_ATTRIBUTES>
<DATASET_ATTRIBUTE>
<TAG>TO DO: Submitter defined tag name</TAG>
<VALUE>TO DO: Submitter defined value</VALUE>
<UNITS>TO DO: Submitter defined units</UNITS>
</DATASET_ATTRIBUTE>
</DATASET_ATTRIBUTES>
</DATASET>
<!--
If you are submitting more than one dataset, replicate the block <DATASET> to </DATASET> here,
as many times as necessary.
-->
</DATASETS>
<!--
Controlled terms for Dataset type:
Whole genome sequencing
Exome sequencing
Genotyping by array
Transcriptome profiling by high-throughput sequencing
Transcriptome profiling by array
Amplicon sequencing
Methylation binding domain sequencing
Methylation profiling by high-throughput sequencing
Phenotype information
Study summary information
Genomic variant calling
-->
STUDY XML file
<?xml version="1.0" encoding="UTF-8"?>
<STUDY_SET>
<STUDY alias="TODO: UNIQUE NAME FOR SUBMISSION (eg. Pancreatic Cancer Genome Sequencing)" center_name="TODO: center name/abbreviation found at http://docs.icgc.org/submission/institutions/ (eg. OICR)">
<DESCRIPTOR>
<STUDY_TITLE>TODO: Title of publication</STUDY_TITLE>
<STUDY_TYPE existing_study_type="TODO: CONTROLLED VOCABULARY END OF XML eg. Whole Genome Sequencing"/>
<STUDY_ABSTRACT>
TODO: STUDY ABSTRACT AS IT COULD APPEAR IN A PUBLICATION
</STUDY_ABSTRACT>
<CENTER_PROJECT_NAME>TODO: Project Name (eg. Pancreatic Cancer Sequencing Initiative)</CENTER_PROJECT_NAME>
</DESCRIPTOR>
<STUDY_ATTRIBUTES>
<STUDY_ATTRIBUTE>
<TAG>Consortium</TAG>
<VALUE>International Cancer Genome Consortium</VALUE>
</STUDY_ATTRIBUTE>
<STUDY_ATTRIBUTE>
<TAG>Consortium Project Name</TAG>
<VALUE>TODO: TAG VALUE MUST CORRESPOND TO Project Name listed in http://docs.icgc.org/submission/projects/</VALUE>
</STUDY_ATTRIBUTE>
<STUDY_ATTRIBUTE>
<TAG>Consortium Project Code</TAG>
<VALUE>TODO: TAG VALUE MUST CORRESPOND TO ICGC PROJECT CODE (eg. PACA-CA). DCC Project codes can be found at http://docs.icgc.org/submission/projects/</VALUE>
</STUDY_ATTRIBUTE>
</STUDY_ATTRIBUTES>
</STUDY>
</STUDY_SET>
<!--
Controlled vocabulary for existing_study_type:
Whole Genome Sequencing
Metagenomics
Transcriptome Analysis
Resequencing
Epigenetics
Synthetic Genomics
Forensic or Paleo-genomics
Gene Regulation Study
Cancer Genomics
Population Genomics
RNASeq
Exome Sequencing
Pooled Clone Sequencing
Other
If using "Other" please add new_study_type="TODO: add own term" attribute
-->
SAMPLE, EXPERIMENT and RUN XML files
For SAMPLE, EXPERIMENT and RUN metadata, only fragments of the XML files are provided to illustrate how certain IDs are referenced across files.
SAMPLE XML file
NOTE: The sample ID specified in the SAMPLE alias field MUST match the DCC-submitted analyzed_sample_id
| Name | VALUE |
|---|---|
| SAMPLE alias | DCC submitted analyzed_sample_id |
| icgc_project_code | ICGC Project code http://docs.icgc.org/submission/projects/ |
| submitter_donor_id | DCC submitted donor_id |
| submitter_specimen_id | DCC submitted specimen_id |
| specimen_type | DCC submitted specimen_type |
| submitter_sample_id | DCC submitted analyzed_sample_id or matched_sample_id |
| gender | DCC submitted donor_sex |
Fragment of the SAMPLE XML file
<SAMPLE_SET xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:noNamespaceSchemaLocation="ftp://ftp.sra.ebi.ac.uk/meta/xsd/sra_1_5/SRA.sample.xsd">
<SAMPLE alias="MUST MATCH DCC-submitted analyzed_sample_id" center_name="TODO: Enter centre name/abbreviation as specified at http://docs.icgc.org/submission/institutions/">
<TITLE>
TODO: A SHORT INFORMATIVE DESCRIPTION OF THE SAMPLE
</TITLE>
<SAMPLE_NAME>
<TAXON_ID>
TODO: PROVIDE NCBI TAXID FOR ORGANISM (e.g. 9606 for human)
</TAXON_ID>
<SCIENTIFIC_NAME>
TODO: SCIENTIFIC NAME AS APPEARS IN NCBI TAXONOMY FOR THE TAXON_ID (e.g. homo sapiens)
</SCIENTIFIC_NAME>
<COMMON_NAME>
TODO: OPTIONAL COMMON NAME AS APPEARS IN NCBI TAXONOMY FOR THE TAXON_ID (e.g. human)
</COMMON_NAME>
</SAMPLE_NAME>
<DESCRIPTION>
TODO: A LONGER DESCRIPTION OF SAMPLE AND HOW IT DIFFERS FROM OTHER SAMPLES
</DESCRIPTION>
<SAMPLE_ATTRIBUTES>
<!--
EGA requires Sample attributes Donor ID, Gender and phenotype for each sample
-->
<SAMPLE_ATTRIBUTE>
<TAG>subject_id</TAG>
<VALUE>project or DCC specific donor_id</VALUE>
</SAMPLE_ATTRIBUTE>
<SAMPLE_ATTRIBUTE>
<TAG>gender</TAG>
<VALUE>DCC-SUBMITTED donor_sex</VALUE>
</SAMPLE_ATTRIBUTE>
<SAMPLE_ATTRIBUTE>
<TAG>phenotype</TAG>
<VALUE>TODO: TAG VALUE, Experimental Ontology Factor (EFO) accession term recommended</VALUE>
<!-- http://www.ebi.ac.uk/efo/ . -->
</SAMPLE_ATTRIBUTE>
<!--
The following sample attributes must have values that are consistent with DCC:
1. icgc_project_code
2. submitter_donor_id
3. submitter_specimen_id
4. specimen_type
5. submitter_sample_id
-->
<SAMPLE_ATTRIBUTE>
<TAG>icgc_project_code</TAG>
<VALUE>TODO: TAG VALUE MUST CORRESPOND TO ICGC PROJECT CODE (eg. PACA-CA). DCC Project codes can be found at http://docs.icgc.org/submission/projects/<VALUES>
</SAMPLE_ATTRIBUTE>
<SAMPLE_ATTRIBUTE>
<TAG>submitter_donor_id</TAG>
<VALUE>DCC-submitted donor_id</VALUE>
</SAMPLE_ATTRIBUTE>
<SAMPLE_ATTRIBUTE>
<TAG>submitter_specimen_id</TAG>
<VALUE>DCC-submitted specimen_id</VALUE>
</SAMPLE_ATTRIBUTE>
<SAMPLE_ATTRIBUTE>
<TAG>specimen_type</TAG>
<VALUE>DCC-submitted specimen_type (eg. Primary tumour - solid tissue)</VALUE>
</SAMPLE_ATTRIBUTE>
<SAMPLE_ATTRIBUTE>
<TAG>submitter_sample_id</TAG>
<VALUE>DCC-submitted analyzed_sample_id or DCC-submitted matched_sample_id</VALUE>
</SAMPLE_ATTRIBUTE>
</SAMPLE_ATTRIBUTES>
</SAMPLE>
</SAMPLE_SET>
Fragment of the EXPERIMENT XML file
<EXPERIMENT_SET>
<EXPERIMENT alias="EXP12345" … >
<STUDY_REF refname="Pancreatic Cancer Genome Sequencing" />
.......
<DESIGN>
<SAMPLE_DESCRIPTOR refname="TODO: SHOULD MATCH DCC-SUBMITTED analyzed_sample_id" />
.......
<LIBRARY_DESCRIPTOR>
<LIBRARY_STRATEGY>
TODO: MUST MATCH DCC-SUBMITTED sequencing_strategy field
</LIBRARY_STRATEGY>
Fragment of the RUN XML file
<RUN alias="SC_RUN_4000_2"/>
<EXPERIMENT_REF refname="EXP12345"/>
<DATA_BLOCK>
<FILES>
<FILE filename="CLLS0001.bam" filetype="bam" />
</FILES>
</DATA_BLOCK>